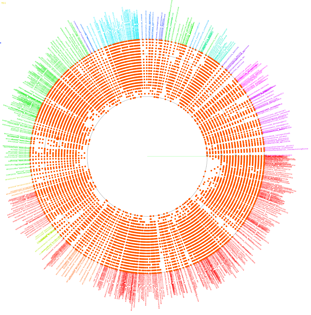
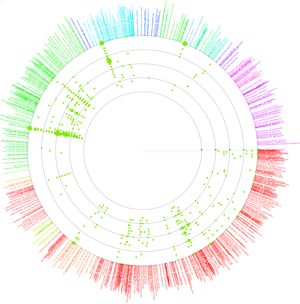
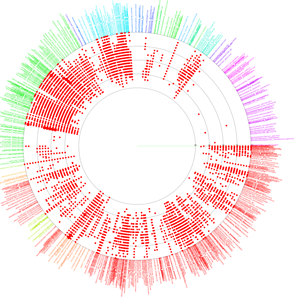
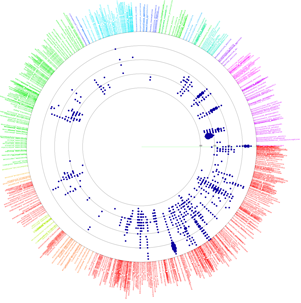
VITCOMIC is a VIsualization tool for Taxonomic COmpositions of MIcrobial Community that can analyze millions of bacterial 16S rRNA gene sequences and calculate the overall taxonomic composition for a microbial community.
The 16S rRNA gene sequences of genome-sequenced species were used as references to identify the nearest relative of each sample sequence. VITCOMIC plots all sequences in a single figure and indicates relative evolutionary distances. VITCOMIC yields a clear representation of the overall taxonomic composition of each sample and facilitates an intuitive understanding of differences in community structure between samples. If you want to know detailed descriptions of the VITCOMIC algorithm, please read our paper.
Try VITCOMIC
Input data:File format1: FASTA file (less than 100,000 sequences) BLASTed file (-m 8)
File format2: flat file .gz .zip
Tag: (use [A-Za-z0-9-_])
Email:
Color of dots: Red Orange Green Blue
How to use
- 1. Input data
- Both of a BLAST result and a Multi-FASTA file are acceptable for the input data in VITCOMIC. The BLAST result is the result of BLASTN against our reference 16S rRNA gene sequence database with -m 8 options. The Multi-FASTA file is the "unaligned" set of 16S rRNA gene sequences obtained from your sample. In case of the Multi-FASTA file, we conduct the BLASTN search against our reference 16S rRNA gene sequence database with -m 8 -e 1e-8 -F F -b 10 -v 10 options before constructing the plot. Since our server has small number of cpu, in case of the Multi-FASTA file, the file that contain less than 100,000 sequences is currently acceptable. Both of the BLAST result and the Multi-FASTA file, the file that is smaller than 1GB is acceptable because of difficulties of the file transfer.
Here is the Sample data of BLAST result and FASTA file obtained and calculated from (Grice EA. et al. 2008, Genome Res.) - 2. File format1
- File format1 is a file format identifier of your upload file. You need to choose the corresponding identifier with your file format. If you want to upload a multi-FASTA file, you should choose FASTA file. If you want to upload a BLAST result against our reference database, you should choose BLASTed file.
- 3. File format2
- File format2 is also a file format identifier of your upload file. To reduce the size of your upload file, we strongly recommend that you compress your file with gzip or zip format. If you don't compress your file, please choose "flat file".
- 4. Tag
- Tag is a identifier of your upload file. Please input any alfabet and number such as "Sample1".
- 5. Email
- Please input your Email address. This Email address is very important because the URL of your VITCOMIC result is send by the Email after calculation (usually less than one day).
- 6. Color of dots
- 7. Output files
- There are three output files that are provided from VITCOMIC.
*.ps
A Postscript file that is the main result of VITCOMIC program. The order of reference species in the plot is described this. Detailed description of the plot is described in our paper.
*.cluster
A text file that describes the taxonomic composition of the sample.
*.query_hit
A text file that decribes the result of taxonomic assignment of each sequence. Each row describes
"the_query_sequence_name" "assigned reference species name" "average similarity".
This file contains only taxonomically assigned query sequences in VITCOMIC.
Reference
Mori H., Maruyama F. and Kurokawa K., VITCOMIC: visualization tool for taxonomic compositions of microbial communities based on 16S rRNA gene sequences. BMC Bioinformatics 2010, 11:332.Journal Link.
Acknowledgement
This work has been supported by BIRD of Japan Science and Technology Agency (JST).